The validation of diagnostic tools to monitor the drug efficacy and the spread of anthelmintic resistance
Scope
Within this work package, we will validate (i) an automated system for quality-assured egg counting containing an integrated system to analyse and report drug efficacy data and (ii) molecular markers for anthelmintic resistance (AR) in human soil-transmitted helminths (STHs).
1. Validation of an automated system for quality-assured egg counting containing an integrated system to analyse and report drug efficacy data.
Recently, TechionGroup has launched the FECPAKG2 as a complete remote-location worm assessment tool for sheep/cattle farmers and their veterinarians to monitor both the intensity of worm infections and the efficacy of administered drugs.
The most important features of FECPAKG2 and their impact on worm diagnosis are illustrated in the table below:
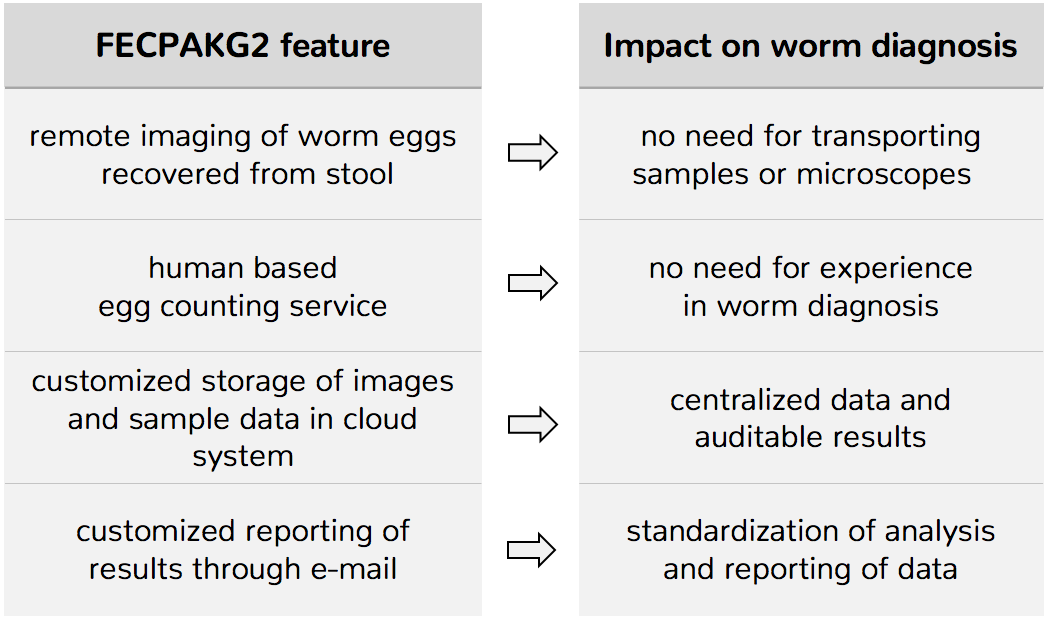
Given the obvious similarities in the morphology of worm eggs, as well as the diagnosis and the study design for assessing drug efficacy, FECPAKG2 is also a promising tool for worm assessment in public health settings.
To automate the detection of eggs in the images produced by the FECPAKG2, we will link the current FECPAKG2 platform with HEAD (Helminth Egg Automatic Detector) software that identifies and quantifies helminth eggs. This software was developed and validated for the detection of helminth eggs in environmental samples by The National Autonomous University of Mexico (UNAM).
Aspects that will further be addressed during this WP are
- the diagnostic performance of FECPAKG2 compared to Kato-Katz, Mini-FLOTAC and qPCR
- the ability of FECPAKG2 to estimate drug efficacy against human STH compared to Kato-Katz, Mini-FLOTAC and qPCR
- the adaptation of the FECPAKG2 software to a public health perspective
- the possibility to automate egg counting, and
- the cost-effectiveness of an automated system for quality-assured egg counting with a built-in analysis compared to the traditional process of stool examination, manual data entry, analysis and reporting
2. Validation of molecular markers for anthelmintic resistance in STHs
Due to unfortunate and costly experiences in veterinary medicine, AR has been researched in detail in animal STHs, providing strong evidence of molecular markers associated with resistance to benzimidazole drugs, such as albendazole and mebendazole. Generally, benzimidazole resistance in animal STHs is caused by single nucleotide polymorphisms (SNPs) in the gene encoding β-tubulin. Various methodologies to detect and quantify these SNPs in animal STHs have been developed, some of which have now also been tested and applied for human STHs in well-equipped laboratories. The few studies that assessed SNPs in human STHs suggest (i) that SNPs are predominantly found in codon 200, (ii) that AR-associated mutations increased after drug administration (T. trichiura), but (iii) that there was no clear connection with the drug efficacy.
In this work package, we will assess:
- the differences in frequency of SNPs linked to AR
- across study sites with a different history of drug administration to assess the long-term impact of drug pressure,
- in responders, poor responders and non-responders to assess the impact of AR on drug efficacy,
- both before and after drug administration to assess the short-term impact of drug pressure;
- the possibility of pooling samples for a resource-saving assessment of SNPs; and
- the performance of loop-mediated isothermal amplification (LAMP) technology as a point-of-care tool for detection of SNPs
Study sites
Trials for this work package will be conducted in Africa (Ethiopia and Tanzania), South America (Brazil) and Asia (Lao PDR). The selection of these sites is based on their experience in assessing drug efficacy and evaluating the performance of diagnostic tools, the availability of well-equipped diagnostic facilities and skilled personnel, and their history of mass drug administration (see figure below).





